Activity lecture location:Home≡Activity lecture≡Details
Professor Sun Liangdan's team mapped the first protein truncation mutation in a Chinese population and revealed its important influence on the pathogenesis of psoriasis
Source:Institute of Dermatology, Anhui Medical University Date:2021/06/22 Browse:1342 second
On June 21, the team of Professor Sun Liangdan from the Department of Dermatology, the First Affiliated Hospital of Anhui Medical University, Anhui Medical University Institute of Dermatology, and the Key Laboratory of Dermatology of the Ministry of Education in the internationally renowned academic journal Genome research ( IF:11.1) published a research paper entitled "Deep sequencing of 1,320 genes reveals the landscape of protein-truncating variants and their contribution to psoriasis in 19,973 Chinese individuals". The Dermatology Department of the First Affiliated Hospital of Anhui Medical University is the first author and first correspondent of this article. The study used in-depth sequencing and analysis of 1,320 genes in nearly 20,000 psoriasis patients and control samples to draw the first large-scale protein truncation mutation map of the Chinese population, and reveal the effect of such mutations on the pathogenesis of psoriasis. The important influence has provided new ideas for the research on the genetic mechanism of complex diseases and therapeutic intervention.

Figure 1. A screenshot of the article homepage
Protein-truncating variants (protein-truncating variant, PTV) refer to a series of severely affected proteins such as termination codons, frameshift or splice site mutations contained in the genome The genetic variation that encodes the integrity and function of genes is of great significance to human phenotypic diversity and the occurrence and development of diseases. PTV may originate from genetic or intergenerational new mutations, and it is widespread in the population. Because the frequency of specific PTV in the population is usually extremely low and has high population specificity, its analysis requires specific large-scale population data and reliable analysis techniques. There is no public PTV map of the Chinese population before this study. .

Figure 2. The protein truncation mutation map on the 1320 gene in this study
This study conducted an in-depth analysis of the PTVs characteristics of 1,320 genes in 10,535 healthy controls and 9,434 patients with psoriasis. A total of 8,720 PTVs were identified. Of these, 77% have never been reported before, showing a high degree of population specificity. Researchers through a new algorithm found that 88% of PTV is harmful and affected by purification selection, showing the shaping and influence of natural selection on human genetic variation. The paper also classified and analyzed all the identified PTVs in detail. Based on this, the first Chinese population protein truncation mutation map was drawn, which perfected the "Chinese puzzle" in this field.
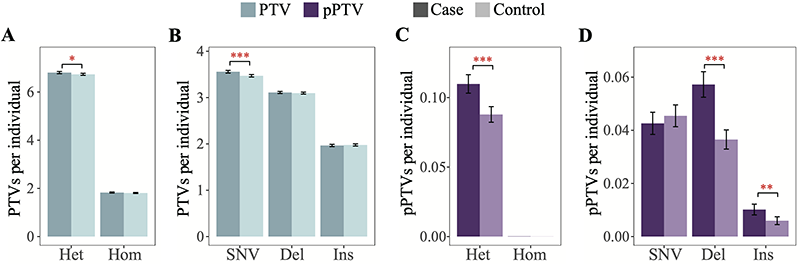
Figure 3. Protein truncation variants are enriched in psoriasis patients
The study found that patients with psoriasis carried more PTV compared with the control group (P=0.02), showing its important influence on the mechanism of disease. The researchers also compared the three major populations of China, Europe, and Africa, and identified 18 PTVs with extremely high levels of differentiation in different populations, and speculated that they may be affected by positive selection, further highlighting the population-specific characteristics of PTV .
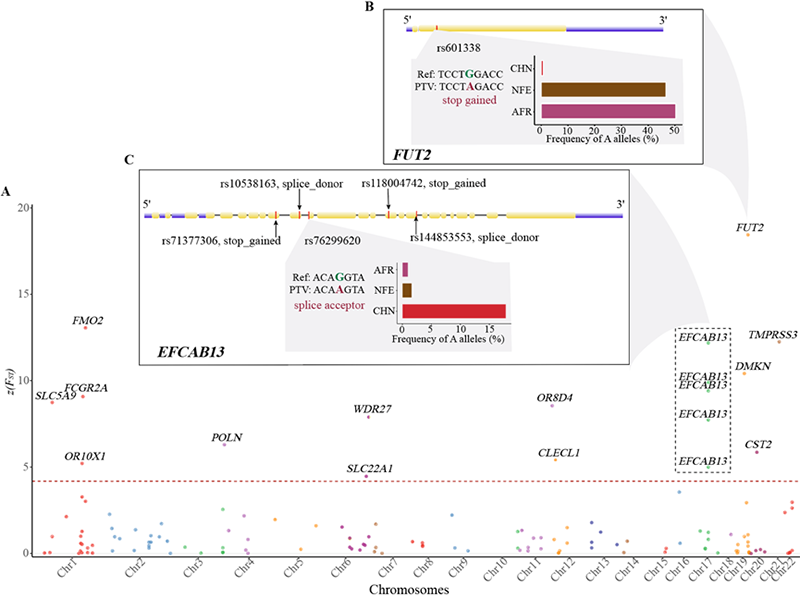
Figure 4. Genes related to protein truncation mutations with highly differentiated allele frequencies in China, Europe, and non-populations
It is reported that doctoral students Xu Huixin, Zhen Qi, bioinformatics experts Bai Mingzhou, Fang Lin and Zhang Yong are the co-first authors. Professor Sun Liangdan from the First Affiliated Hospital of Anhui Medical University is the corresponding author. Researcher Jin Xin from the School of Medicine of South China University of Technology and Professor Joshua M. Akey from Princeton University are the co-corresponding authors. Professor Sun Liangdan’s team has long been committed to the research of disease genetics and translational medicine, analyzing the disease genome variation map, and has successively discovered single nucleotide variation in psoriasis susceptibility genes (Nat Genet, 2009, 2010; Nat Commun, 2014); copy number variation (J Invest Dermatol, 2010); coding variants (Nat Genet, 2014; Nat Commun, 2015; J Invest Dermatol, 2014), HLA variants (Nat Genet, 2016) and indel variants (J Invest Dermatol, 2019), systematic interpretation The genetic susceptibility mechanism and genetic etiology of psoriasis provide a basis for precision medicine research. This study constructs a protein truncation mutation map of the Chinese population, further enriches the basis of psoriasis genetic variation, expands the understanding of the population specificity of protein truncation mutations, and is of great significance for the origin and evolution of the Chinese population and the study of the genetic mechanism of the disease.
Original link:
China Psoriasis Network CPC2020

Copyright: China Psoriasis Network Wanicp Bei 11019793 皖公网安备 34019202000792号